Segminator II
Segminator II has been developed and implemented by
John Archer,
as part of a project funded by the BBSRC (project grant: BB/H012419/1) that is supervised by
David Robertson
and Andrew Rambaut.
It is a software for the characterization of viral read data that is generated on next generation sequencing platforms including:
454 Life Sciences,
Illumina,
Ion Torrent and
Pacific Biosciences.
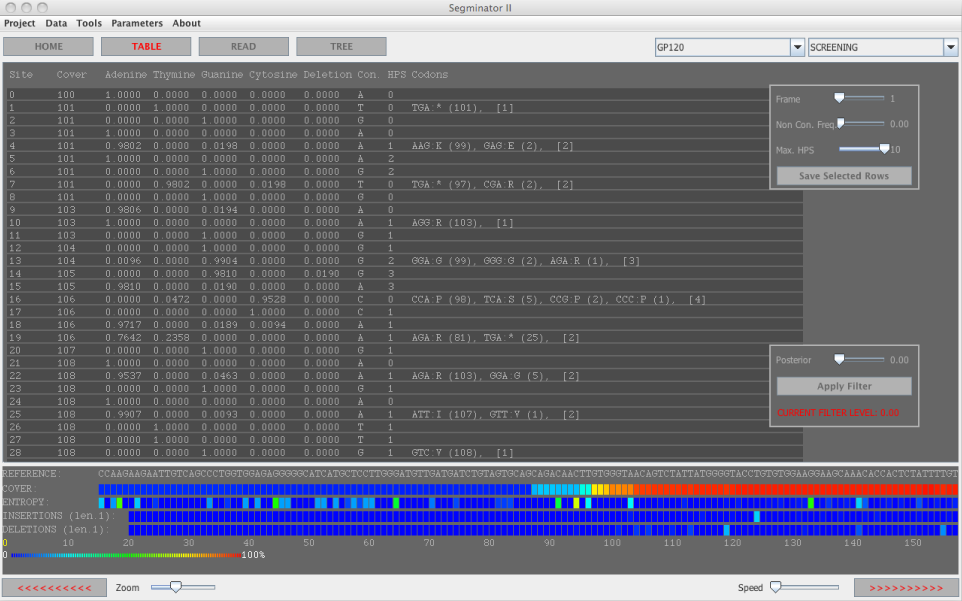
Figure 1: Main display area of Segminator II
Notable features of the software include:

Figure 1: Main display area of Segminator II
Notable features of the software include:
- the seamless mapping and aligning of read data to a reference sequence,
- the association of multiple datasets to a single reference sequence,
- the per site coverage, entropy and base frequencies, along with the ability to query sites based on the latter,
- codon frequencies and consensus sequence generation,
- phylogenetic inference,
- read visualization and,
- a bayesian framework for platform error correction.(new)
free for academics | GNU Lesser GPL licence | software not guaranteed


